Indexing & Abstracting
Full Text
Research ArticleDOI Number : 10.36811/ijbm.2021.110023Article Views : 69Article Downloads : 44
Self-Dividing Micelles: A Mechanistic Look with Evolutionary and Clinical Implications
Bruce K Kowiatek
Blue Ridge Community and Technical College, USA
*Corresponding Author: Bruce K. Kowiatek. Blue Ridge Community and Technical College, 13650 Apple Harvest Dr, Martinsburg, WV 25403, USA. E-mail: bkowiate@blueridgectc.edu
Article Information
Aritcle Type: Research Article
Citation: Bruce K Kowiatek. 2021. Self-Dividing Micelles: A Mechanistic Look with Evolutionary and Clinical Implications. Int J Biol Med. 3: 08-20.
Copyright: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Copyright © 2021; Bruce K Kowiatek
Publication history:
Received date: 26 July, 2021Accepted date: 03 August, 2021
Published date: 05 August, 2021
Abstract
Micellar therapy has become a usefully viable treatment arm in various fields, ranging from oncology to bioimaging. As such, research leading to any improvements or adaptations in administration and techniques can have far-reaching consequences. Potential aspects of prebiotic chemistry may also be explored in such research as well. To that end, proof-of-concept experiments were performed to elucidate a possible mechanism of action for prebiotic protocell division. Representative potentially prebiotically plausible biomolecules, i.e., a fatty acid, amino acid, and nucleotide were mixed and heated in water and subjected to microscopic examination for observation of possible self-division and laboratory testing for the presence of polypeptides and polynucleotides (Biuret, MALDI mass-spec, etc.) with and without the presence of nucleotide. The results are presented for the first time here and a mechanism is proposed that best fits the data obtained. The evolutionary, e.g., prebiotic biomolecular cooperativity, and clinical, e.g., potential antineoplastic micellar/vesicular therapy, ramifications are discussed as well.
Keywords: Micelle; Liposome; Protocell; MRNA; Self-division; Mechanism; Solid tumors
Introduction
Micellar therapy has become a usefully viable treatment arm in various fields, ranging from oncology to bioimaging [1]. As such, research leading to any improvements or adaptations in administration and techniques can have far-reaching consequences. Potential aspects of prebiotic chemistry may also be explored in such research as well. To that end, proof-of-concept experiments were performed to elucidate a possible mechanism of action for prebiotic protocell division. Although successful pioneering research in this area has been previously performed by Dreamer [2] and Szostak [3] among others, to this author’s knowledge none has satisfactorily addressed 1) the dependence upon nucleic acid for protocell self-division, 2) the resultant production of other prebiotically relevant biomolecules, i.e., ATP, that further promulgate the self-division process, and 3) the recycling of the process to promulgate further self-divisions. This paper addresses all three. The results are presented for the first time here and a mechanism is proposed that best fits the data obtained. The evolutionary, e.g., prebiotic biomolecular cooperativity, and clinical, e.g., potential antineoplastic micellar/vesicular therapy, ramifications are discussed as well.
Experimental
All research was conducted at Blue Ridge Community and Technical College in Martinsburg, WV, USA from April of 2018 through January of 2019. All pH measurements were made using a Horiba TwinpH waterproof B-213 Compact pH meter. Glass microscope well slides were procured through American Science and Surplus. Zeiss Primo Star binocular microscope and corresponding software equipment was used for imaging. All chemical supplies were purchased from Fisher Scientific. All Pyrex glassware was sterilized at 130° C for one hour [4] via autoclave using a Quincy Lab Inc. Model 30 GC Lab Oven. All measurements of chemicals were standardized to 0.1 Molarity (M) ± 5% using an Ohaus Analytical Plus electronic balance accurate to within ± 0.0001 gram (g). Three trials per step were performed and recorded with the data presented here representing the average of that total data. All data collected fell within a statistically acceptable ± 5% (p=0.05) internal margin of variance [5] with no outliers using Microsoft Excel Analyze-it add-in software. Concentrations of chemicals used were sufficient to achieve critical micelle concentration (CMC) as per Fisher Scientific product information. To 30 milliliters (mL) of sterile deionized water adjusted to a pH of 7.0 via dropwise titration with 0.1 M hydrochloric acid (HCl) and 0.1 M sodium hydroxide (NaOH) to facilitate imidazolium ion formation in L-histidine (His) [6] was added 0.3 gram (g) of n-Decyl-β-D-malt side and 0.1 g of His and heated to 100° Celsius (C) for 10 minutes (min) while stirred with a magnetic stirring rod. Samples were procured via dropper and subjected to a Biuret Test for polypeptides, with additional samples added to microscope well slides with methylene blue dye added to aid microscopic visualization. Observations were made at 100x oil immersion magnification and recorded. 0.1 g of adenosine 5?-monophosphate (AMP) sodium (Figure 1) was then added to the original mixture and heated to 100° C for 10 min while stirred with a magnetic stirring rod. Samples were procured via dropper and added to microscope well slides with methylene blue dye added. Observations were then made at 100x oil immersion magnification and recorded.
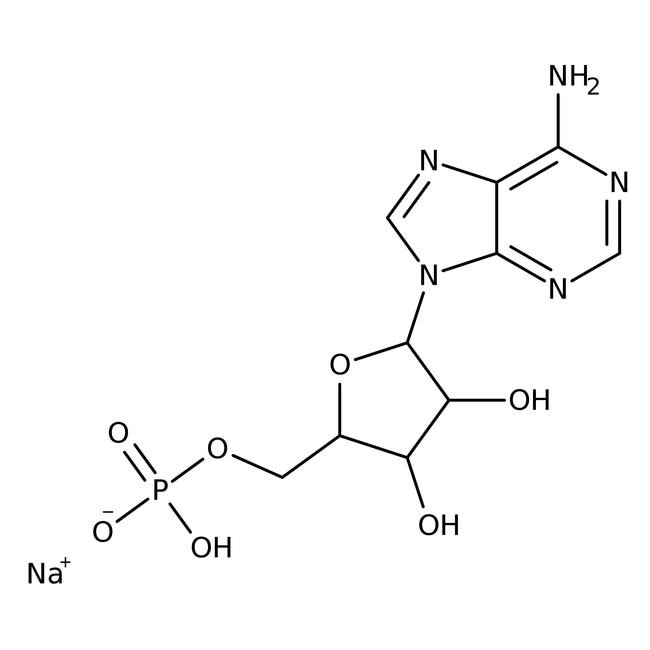
Figure 1: Adenosine 5?-monophosphate (AMP) sodium.
Results and Discussion
Samples sans dye without AMP pre- and post-heating were subjected to a Biuret Test [7] in sterile test tubes and revealed the following:

Figure 2: Pre-heating negative Biuret Test (left) of n-Decyl-β-D-malt side/His sample and post-heating positive Biuret Test (right) of n-Decyl-β-D-malt side/His sample indicating synthesis of poly-L-histidine peptide. Additional samples sans dye was mailed via USPS Priority Mail to Michigan State University (MSU) Mass Spectrometry and Metabolomics Core in East Lansing, MI, USA for analysis and the results emailed back. Microscopic observation of the first set of samples without AMP revealed the following.
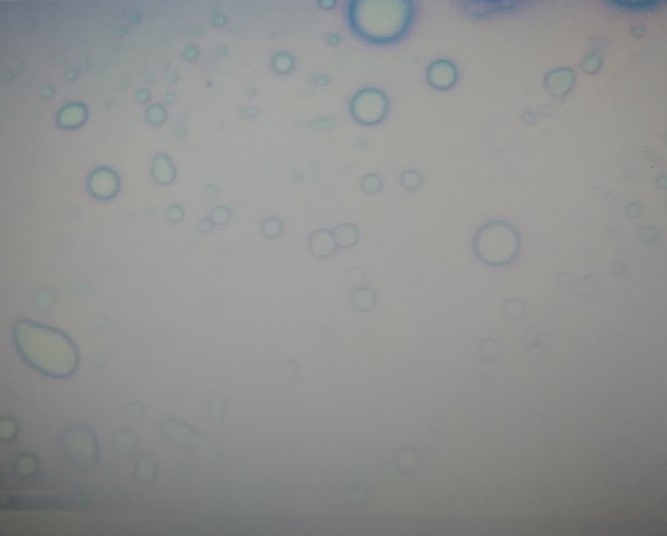
Figure 3: 100x magnification oil immersion micrograph of n-Decyl-β-D-malt side/His micelles (~1.0 µm) with methylene blue dye. Microscopic observation of the second set of samples with AMP revealed.

Figure 4: 00x magnification oil immersion micrograph of n-Decyl-β-D-malt side/His/AMP individual micelle (~ 0.5 µm) pre-division with methylene blue dye.

Figure 5: 100x magnification oil immersion micrograph of n-Decyl-β-D-malt side/His/AMP individual micelle during division with methylene blue dye.

Figure 6: 100x magnification oil immersion micrograph of n-Decyl-β-D-malt side/His/AMP individual micelle post-division with methylene blue dye.
Full 1:30 recording of representative micelle divisions can be found at https://www.youtube.com/watch?v=2zjDAdGUeZM
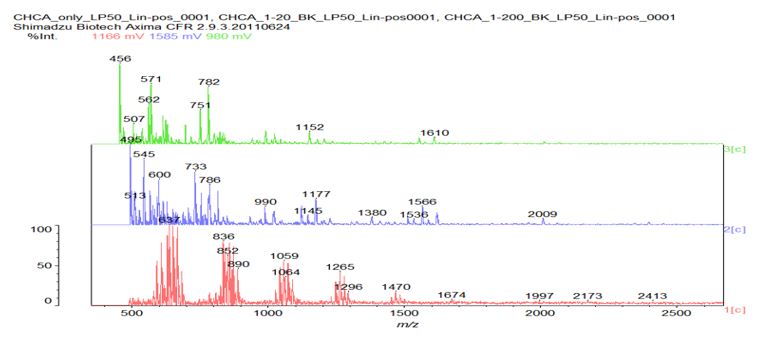
Figure 7: Matrix-assisted laser desorption / ionization (MALDI) mass spectrograph of a representative n-Decyl-β-D-maltoside/His/AMP sample sans dye. Bottom (red) line indicates α-Cyano-4-hydroxycinnamic acid (CHCA) matrix only; middle (blue) line indicates CHCA matrix plus sample at 1/20 dilution; top (green) line indicates CHCA matrix plus sample at 1/200 dilution. Note peak at 507 m/z in top line and peaks at 990 and 1380 m/z respectively in middle line. No direct micelle division was microscopically observed in the post-heating n-Decyl-β-D-malt side/His samples; however, direct micelle division was microscopically observed in all of the post-heating n-Decyl-β-D-malt side/His/AMP samples, indicating that the presence of AMP triggered the micelle division. N-Decyl-β-D-malt side was chosen as a representative fatty acid-like nonionic detergent amenable to MALDI spectroscopy (Fisher Scientific product information); His was chosen as a representative prebiotically plausible amino acid [8] and due to its dual polymerizing and cleaving capacity as a polyimide [9]; AMP was chosen as a representative prebiotically plausible nucleotide [10] and due to its being the direct precursor of adenosine 5?-triphosphate (ATP) [11] (Figure 8).
Figure 8: Adenosine 5?-triphosphate (ATP).
Heating was applied to induce amino acid and nucleotide polymerization via condensation dehydration reactions and appears to have been successful as evidenced by positive Biuret tests and MALDI spectroscopy peaks in the blue 1/20 dilution middle line of Figure 7 at 1380 and 990 m/z, respectively indicating a 10-mer polymer decapeptide of protonated poly-L-histidine imidazolium ions [12] and a 3-mer polymer trinucleotide of protonated polyadenylate [13]. Interestingly, the peak at 507 m/z in the green 1/200 dilution top line indicates the presence of unhydrolyzed ATP [14] and suggests a possible mechanism for not only the robust polymerization of short nucleotides that mimics nature’s own, but for micelle self-division as well. The mechanism proposed here begins with the heat-induced condensation dehydration polymerization of individual His molecules into 10-mer protonated imidazolium ion (Figure 9) poly-L-histidine decapeptides (Figure 10), present in the micelles pictured in Figure 3, with the partially negative protonated hydroxy dipole moiety of malt side on the surface of the micelle forming an electrostatic or possibly even hydrogen bond with the partially positive protonated amino dipole moiety adjacent to the α-carbon of the poly-L-histidine (Figure 11).
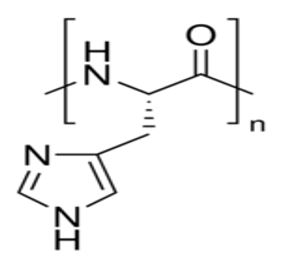
Figure 9:
Figure 10: Poly-L-histidine decapeptide (n = 10).
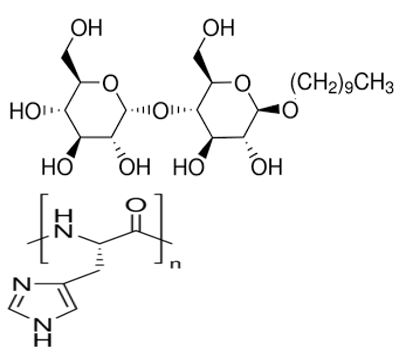
Figure 11: Proposed surface structure of n-Decyl-β-D-malt side/poly-L-histidine micelle.
With the addition of AMP, the negatively charged oxygens of the molecule’s phosphate moieties align with the positively charged imidazolium ions of the poly-L-histidine decapeptide (Figure 12) to accommodate at least nine AMPs per poly-L-histidine decapeptide which would facilitate, accompanied by the hydrolysis of six adenosine moieties out of every nine AMPs, the formation of three ATP molecules. Such accommodation of at least nine AMPs and subsequently three ATPs would mimic nature and facilitate the formation of polyadenylate trinucleotide accompanied by the hydrolysis of three inorganic pyrophosphate (PPi) molecules, inducing the momentary inversion of the micelle that would facilitate both release of the ATP and polyadenylate trinucleotide respectively, and ultimately self-division of the micelle (Figure 13). The detection of any adenosine moieties and PPi molecules fell below the limits of the MALDI analysis performed here.
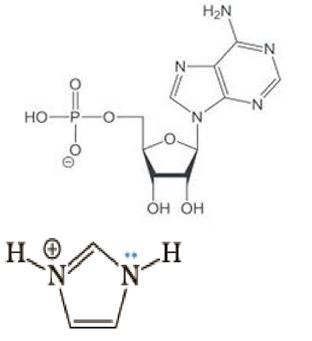
Figure 12: Alignment of negatively charged oxygen on AMP with positively charged imidazolium ion.
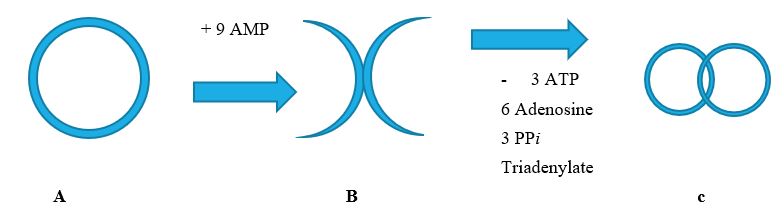
Figure 13: A: n-Decyl-β-D-malt side/poly-L-histidine micelle heated to 100° C (outer surface proposed structure pictured in Figure 11) that, with the addition of at least 9 AMP molecules, inverts the micelle (B) (inverted surface proposed structure pictured in Figure 14), polymerizing the AMP and hydrolyzing 6 adenosine moieties, forming 3 ATP molecules which subsequently hydrolyze 3 PPi, synthesizing the 3-mer trinucleotide triadenylate, and forms two “daughter” micelles (C), both now with the same outer surface structure as A.
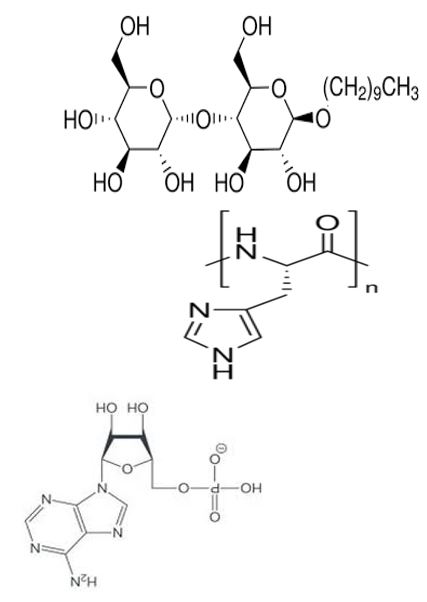
Figure 14: Proposed structure of inverted n-Decyl-β-D-maltoside/poly-L-histidine/polyadenylate micelle membrane.
Evolutionary Implications
As previouosly discussed, all materials used except for the fatty acid-like n-Decyl-β-D-maltoside are prebiotically plausible molecules. Fatty acids themselves, however, are indeed prebiotically plausible molecules [15], and to that end were also employed in these experiments using mono- and polyunsaturated fatty acids, L-lysine, and AMP (see Supplemental Figures S1-S4), as well as n-Decyl-β-D-maltoside, L-lysine, and AMP (see Supplemental Figures S5-S9) with mixed results. Interestingly in all cases though, similar self-dividing protocell vesicle/micelle interactions were observed microscopically with both monolayer and bilayer surfaces, pointing to the reasonable conclusion that life likely arose via inclusive cooperation among prebiotically present biomolecules such as fatty acids, amino acids, and nucleotides under condensing dehydrating conditions rather than the more exclusive prevailing theories like the RNA World Hypothesis. Also interesting is the preferential synthesis of triplet trinucleotides, implicated in everything from the possible origins of metabolism [16], through triplet nucleotide sequences having been shown experimentally to act as templates for RNA-catalyzed RNA ribozymes when nucleotide triphosphates were used as substrates [17], to the highly likely 93-nucleotide primordial precursor transfer RNA (tRNA) molecule proposed by Burton et al [18].
Clinical Implications and Prospectus
One of the clinical implications of either self-dividing monolayer micelles or bilayer liposomes is arguably obvious, with the former delivering lipophilic chemotherapeutic antineoplastic agents and the latter delivering hydrophilic ones, offering a distinct advantage over their current non-dividing counterparts presently in use, providing significantly improved coverage over a much larger surface area, particularly in treating solid tumors. The choice of polypeptide employed may also not need be limited to poly-L-histidine but can also be expanded to include any of the basic side-chain amino acids, such as the now-predominant poly-L-lysine, as well as possibly poly-L-arginine. Another not as readily apparent implication involves the potential clinical application of the triplet trinucleotides themselves, specifically the possible use of messenger RNA (mRNA) stop codons. With the highly touted mRNA technology currently being successfully incorporated into several of the SARS-CoV-2 (Covid-19) vaccines now in production, the prospect of the use of this technology in potential cancer vaccines is presently in various stages of implementation [19]. The proposition is therefore thus put forth here for the possible development of an mRNA cancer vaccine or vaccines utilizing self-dividing micelles and/or liposomes using only stop codons for the treatment and destruction of rapidly-dividing solid tumors, after which time clinical trials assessing safety and efficacy would be justified.
Acknowledgements
I especially thank my loving family for their generous gift of time in the performance of these experiments and the writing of this paper, as well as my fellow faculty and staff at Blue Ridge Community and Technical College for the generous use of their facilities and equipment. Very special thanks also to Dr. Zachary Burton, Professor Emeritus at Michigan State University (MSU) for invaluable insight, discussion, and financial support, without whom this paper would not have been possible, as well as the Michigan State University (MSU) Mass Spectrometry and Metabolomics Core in East Lansing, MI, USA.
Abbreviations
AMP: adenosine monophosphate; ATP: adenosine triphosphate; His: L-histidine; MALDI: Matrix-assisted laser desorption/ionization; mRNA: messenger ribonucleic acid; tRNA: transfer ribonucleic acid

Figure S1: 100x magnification oil immersion methylene blue-stained micrograph of self-dividing fatty acid/poly-L-lysine/polyadenylate vesicles.
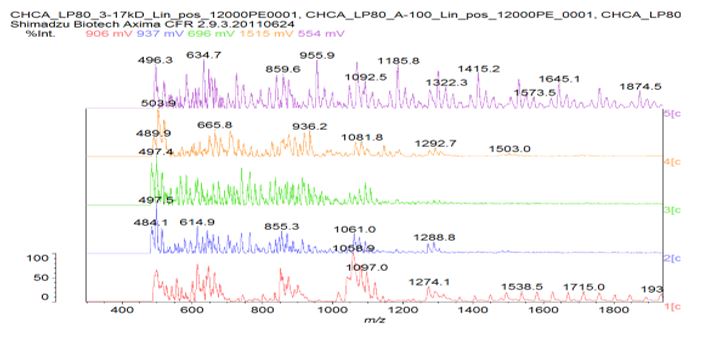
Figure S2: MALDI analysis of sample A compared to a poly-asparagine standard from Sigma. Used CHCA matrix and tried different dilutions of sample A.
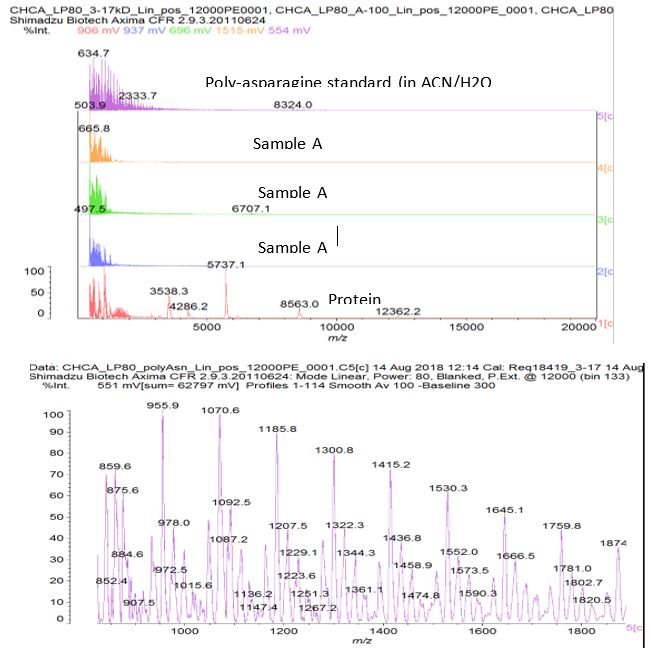
Figure S3: Zoom in to show poly-asparagine standard – peaks separated by mass ~114 which is the residue mass of asparagine (asparagine can also deaminate to aspartate which has a residue mass of 115). Poly-lysine should have a repeating unit of mass 128. There is not a clear indication of this in the ‘A’ sample dilutions that were tried.
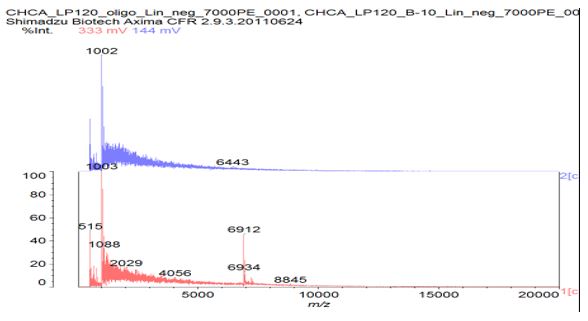
Figure S4: Prepared as last time, using a ZipTipC18 pipette tip to desalt the samples. Could detect the oligonucleotide standard but could not see any obvious signal for sample B in negative ion mode.

Figure S5: 100x magnification oil immersion methylene blue-stained micrograph of self-dividing n-Decyl-β-D-maltoside /poly-L-lysine/polyadenylate vesicles.
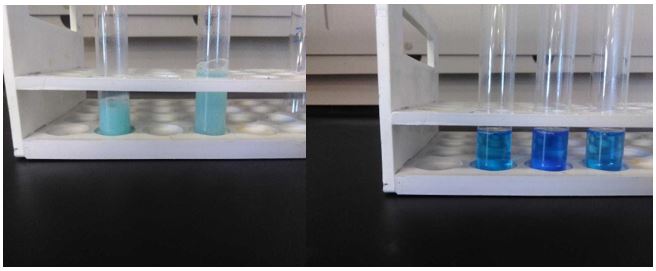
Figure S6: From left to right: negative Biuret tests for n-Decyl-β-D-maltoside in sterile deionized water at room temperature and heated to 100° C; minimally positive Biuret test for poly-L-lysine in n-Decyl-β-D-maltoside/Lys, highly positive Biuret test for poly-L-lysine in n-Decyl-β-D-maltoside heated to 100° C, and another minimally positive Biuret test after addition of AMP.
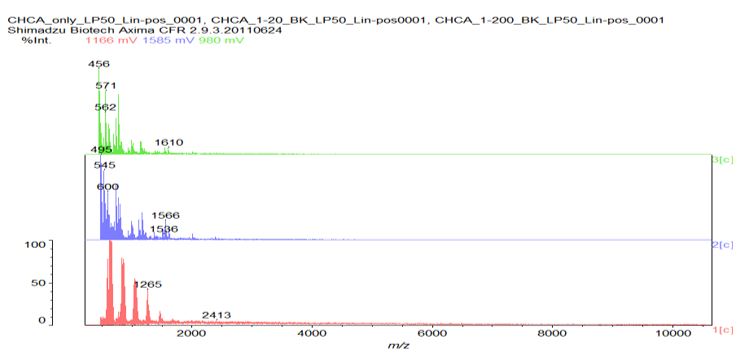
Figure S7: MALDI spectrograph of n-Decyl-β-D-maltoside/Ly’s post heating. Note peak at 1536 m/z indicating synthesis of 12-mer poly-L-lysine dodecapeptide.
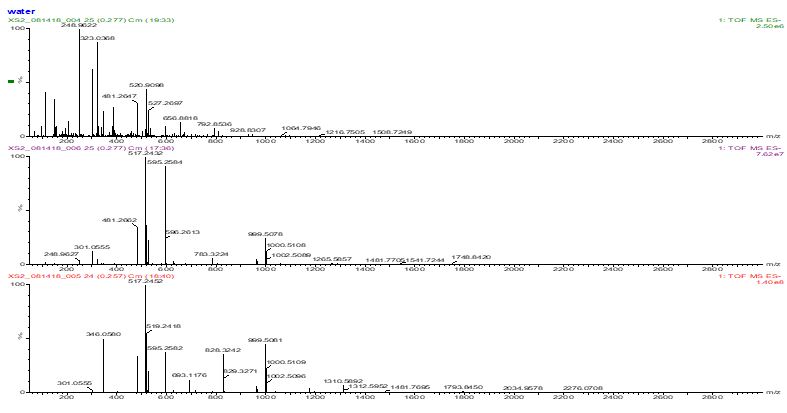
Figure S8: Ran samples by flow-injection on a QTof LC/MS instrument using positive and negative ion electrospray ionization.
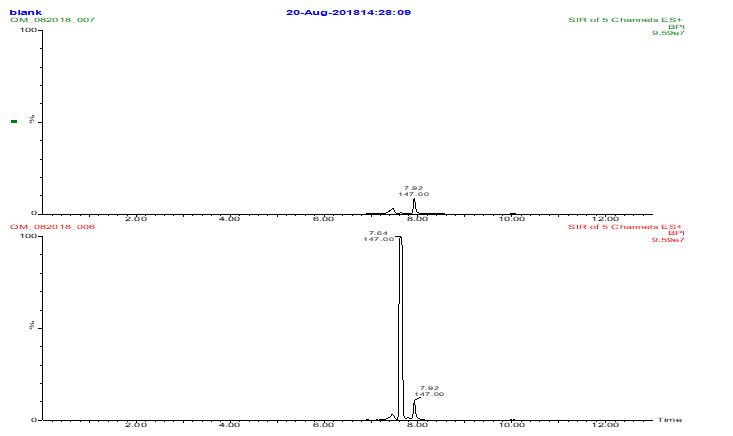
Figure S9: Diluted sample A and analyzed by LCMS using a reversed phase C18 column with an ion-pairing (perfluoroheptanoic acid) containing mobile phase to get lysine retained. The MS was set up to look for lysine (m/z 147) and several polylysine masses (275, 403, 531, and 659). Only a lysine peak was seen. Scanning up to higher mass (m/z 1500) did not reveal anything either.
References
1. Andrade F, Almeida A, Rafael D, et al. 2018. Micellar-based nanoparticles for cancer therapy and bioimaging. Nanooncology. 5: 211-238.
2. Deamer D. 2017. The Role of Lipid Membranes in Life’s Origin. Life. 7: 5. Ref.: https://pubmed.ncbi.nlm.nih.gov/28106741/ Doi: https://doi.org/10.3390/life7010005
3. Szostak JW, Bartel DP, Luisi PL. 2001. Synthesizing life. Nature. 409: 4. Ref.: https://pubmed.ncbi.nlm.nih.gov/11201752/
4. Black J. 1993. Microbiology. Prentice Hall. 334.
5. Bolton S. 1997. Pharmaceutical statistics (3rd Edn.). New York: Marcel Dekker.
6. Oullette R, Rawn JD. 2014. Introduction to Organic Reaction Mechanisms. Elsevier Organic Chemistry. 1: 75-110.
7. O’Donnell MJ, Kowiatek BK. 2019. General, Organic, & Biological Chemistry Laboratory Manual for CHEM 127 & CHEM 128. Kendall Hunt. 211-212.
8. Shen C, Yang L, Miller SL, et al. 1990. Prebiotic synthesis of histidine. Mol Evol. 31: 167-174. Ref.: https://pubmed.ncbi.nlm.nih.gov/11536478/ Doi: https://doi.org/10.1007/bf02109492
9. Cheng L, Abhilash KG, Breslow R. 2012. Binding and biometric cleavage of the RNA poly(U) by synthetic polyimidazoles. PNAS. 109: 12884-1288. Ref.: https://pubmed.ncbi.nlm.nih.gov/22826260/ Doi: https://doi.org/10.1073/pnas.1210846109
10. Kim H, Benner SA. 2017. Prebiotic stereoselective synthesis of purine and noncanonical pyrimidine nucleotide from nucleobases and phosphorylated carbohydrates. PNAS. 114: 11315-11320. Ref.: https://pubmed.ncbi.nlm.nih.gov/29073050/ Doi: https://doi.org/10.1073/pnas.1710778114
11. Frederich M, Balschi JA. 2002. The relationship between AMP-activated protein kinase activity and AMP concentration in the isolated perfused rat heart. J Biol Chem. 277: 1928-1932. Ref.: https://pubmed.ncbi.nlm.nih.gov/11707445/ Doi: https://doi.org/10.1074/jbc.m107128200
12. Tiengo A, Barbarini N, Troiani S, et al. 2009. A Perl procedure for protein identification by Peptide Mass Fingerprinting. BMC Bioinformatics. 10: 1471-2105. Ref.: https://pubmed.ncbi.nlm.nih.gov/19828071/ Doi: https://doi.org/10.1186/1471-2105-10-s12-s11
13. Grillo MP, Lohr MT. 2009. Covalent Binding of Phenylacetic Acid to Protein in Incubations with Freshly Isolated Rat Hepatocytes. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 37: 1073-1082. Ref.: https://pubmed.ncbi.nlm.nih.gov/19196839/ Doi: https://doi.org/10.1124/dmd.108.026153
14. PubChem Identifier: CID 5957.
15. Lopez A, Fiore M. 2019. Investigating Prebiotic Protocells for a Comprehensive Understanding of the Origins of Life: A Prebiotic Systems Chemistry Perspective. Life (Basel). 9: 49. Ref.: https://pubmed.ncbi.nlm.nih.gov/31181679/ Doi: https://doi.org/10.3390/life9020049
16. Kowiatek BK. 2019. Non-enzymatic Methylation of Cytosine in RNA by S-adenosylmethionine and Implications for the Evolution of Translation. J Biotechnol Biomed. 2: 048-056.
17. Attwater J, Raguram A, Morgunov AS, et al. 2018. Ribozyme-catalysed RNA synthesis using triplet building blocks. Elife 15: 7. Ref.: https://pubmed.ncbi.nlm.nih.gov/29759114/ Doi: https://doi.org/10.7554/elife.35255
18. Kim Y, Kowiatek B, Opron K, et al. 2018. Type-II tRNAs and evolution of translation systems and the genetic code. Int J Mol Sci. 22: 19. Ref.: https://pubmed.ncbi.nlm.nih.gov/30360357/ Doi: https://doi.org/10.3390/ijms19103275
19. Copur MS. 2021. Messenger RNA Vaccines: Beckoning of a New Era in Cancer Immunotherapy. Oncology. 35: 4. Ref.: https://pubmed.ncbi.nlm.nih.gov/33893760/ Doi: https://doi.org/10.46883/onc.2021.3504.0198




















