Indexing & Abstracting
Full Text
Research ArticleDOI Number : 10.36811/ijbm.2019.110002Article Views : 3267Article Downloads : 37
AtCIPK19 aggrandizes polyamines-involved cold stress tolerance in plant cells
Wei Tang1,2* and Wells Thompson2
1College of Horticulture and Gardening, Yangtze University, Jingzhou, Hubei 434025, China
2CIMAS, 101 Science Drive, Duke University, Durham, NC 27708, USA
*Corresponding author: Wei Tang, College of Horticulture and Gardening, Yangtze University, Jingzhou, Hubei 434025, China, Tel: +86-13921074062; Fax: +86-716-8066262; Email: wt10yu604@gmail.com
Article Information
Aritcle Type: Research Article
Citation: Wei Tang, Wells Thompson. 2019. AtCIPK19 aggrandizes polyamines-involved cold stress tolerance in plant cells. Int J Biol Med. 1: 03-21.
Copyright: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Copyright © 2019; Wei Tang
Publication history:
Received date: 19 December, 2018Accepted date: 02 January, 2019
Published date: 03 January, 2019
Abstract: The CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASEs (CIPKs) play important roles in regulating ion homeostasis and stress responses. However, molecular mechanism of the Arabidopsis (Arabidopsis thaliana) CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19 (CIPK19) enhanced cold stress tolerance is not fully understood. Here, we report that overexpression of the Arabidopsis CIPK19 gene (AtCIPK19) results in increasing cell viability and growth rate under cold stress in O. sativa, G. hirsutum, and P. strobus. AtCIPK19 increases cold stress tolerance by decreases the thiobarbituric acid reactive substances (TBARS) and increases the levels of putrescine (Put), spermidine (Spd), and spermine (Spm). In AtCIPK19 transgenic rice cell lines, the transcript levels of genes associated with biosynthesis of Put, Spd, and Spm include OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH, OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4 are increased under cold stress. These results will increase our understanding of AtCIPK19-related cold stress tolerance in different plant species and are valuable in plant molecular breeding application.
Keywords: CALCINEURIN B-LIKE PROTEIN; Cold stress; Pinus; Putrescine; Spermidine; Spermine
Introduction
Cold stress reduces plant growth and crop yield. To counteract the damage caused by cold stress, plants have developed capabilities to adapt to this extreme environment, including activation of hormone signaling and alterations of expression of cold stress-related genes. In order to fully characterize the adaptive responses of plants to cold stress, it is important to determine the final levels of mRNAs and proteins in order to reveal the precise novel regulatory mechanisms which specifically function in the response to cold stress [1]. Transcription factors participate in biological processes of cold stress responses. In Prunus mume, increased expression of 15 putative PmNACs transcription factors was observed to tolerate freezing-stress [2]. In rice, OsmiR156 enhances cold stress tolerance by regulating the expression of transcription factor genes [3]. Membrane-to-nucleus signals modulate plant cold tolerance [4]. A yeast one-hybrid assay demonstrated that AaDREB1 encodes a transcription activator and specifically binds to DRE/CRT to enhance tolerance to low temperature [5]. In cucumber, CsWRKY46 was up-regulated in response to cold stress and its overexpression increased the expression of stress-inducible genes, including RD29A and COR47 [5]. In K. obovata, mRNA expression analysis indicated that the KoHSP70 was increased significantly after 48 h cold stress, and reached the highest level at 168 h after cold treatment [6]. In wheat and barley, high transcription level of dehydrin was observed after plants exposed to cold stress [7]. In Glyycine soja, over-expression of GsZFP1 increased the expression of stress-response marker genes, including CBF1, CBF2, CBF3, NCED3, COR47, and RD29A in response to cold stress [8].
Polyamines including putrescine, spermidine, and spermine might help plants to deal with cold stress by interacting with negatively charged macromolecules and regulate their functions [9]. In apple (Malus domestica), genome-wide investigation showed that 18 S-adenosylmethionine decarboxylase and spermidine synthase-related sequences were involved in polyamines biosynthesis in response to cold stressed condition [10]. In rubber tree, S-Adenosylmethionine decarboxylase (SAMDC), a key rate-limiting enzyme involved in polyamines biosynthesis, plays important roles in cold stress response [11]. In bamboo, the endogenous polyamines significantly increased to avoid injury during cold stress [12]. In trifoliate orange [Poncirus trifoliata (L.) Raf.], miRNA396b (ptr-miR396b) positively regulates cold tolerance through reducing ACO transcript levels and simultaneously promoting polyamine synthesis [13]. In Banana, the chilling tolerance induced by NO treatment might be ascribed to the enhanced catabolism of polyamine [14]. In tea (Camellia sinensis) subjected to low-temperature stress, the CsSPMS gene is quickly induced by cold stress [15]. In rice, SamDC, a key enzyme in the polyamine biosynthesis pathway, functions in response to the abiotic stress treatments of cold [16,17]. In Siberian spruce (Picea obovata), the accumulation of polyamines may act as compatible solutes or cryoprotectants to act on freezing tolerance development [18,19].
Calcineurin B-like protein interacting protein kinases (CIPKs) are vital elements in plant abiotic stress signaling pathways. Calcineurin B-like protein interacting protein kinases (CIPKs) are key regulators of pre-transcriptional and post-translational responses to cold stress. In Arabidopsis thaliana, CIPK16 (AtCIPK16) was identified as a regulator of cold stress tolerance [20]. In roots of rice seedling, the serine/threonine/tyrosine protein kinase gene, OsACTPK1, accumulated under abiotic stress [21]. In Brachypodium distachyon, BdCIPK31 was downregulated by polyethylene glycol, NaCl, H2O2, and abscisic acid (ABA) treatments. Overexpression of BdCIPK31 could elevate several stress-associated gene expressions under stress conditions [22]. In Arabidopsis thaliana, the calcineurin B-like protein-interacting protein kinase-7 (AtCIPK7) could negatively regulate TuYV export from infected cells [23]. In wheat, TaCIPK29 was identified as a regulator of stress tolerance. Over-expression of TaCIPK29 in tobacco resulted in increased salt tolerance [24]. In rice, OsCIPK14 and OsCIPK15, are rapidly induced by stress [25]. In Arabidopsis, the calcineurin B-like protein-interacting protein kinase 9, AtCIPK9, acted as a critical regulator of abiotic stress [26].
It has been reported that the Arabidopsis (Arabidopsis thaliana) CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19 (AtCIPK19) acts as an important element to pollen tube growth [27]. Overexpression of AtCIPK19 was associated with elevated cytosolic Ca2+ throughout the bulging tip, indicating that CIPK19 may be involved in maintaining Ca2+ homeostasis through its potential function in the modulation of Ca2+ influx [28]. Although CIPKs play important role in cold stress tolerance, how AtCIPK19 regulates cell response to cold stress is not fully understood. In this investigation, we demonstrated that AtCIPK19 increases cold stress tolerance in O. sativa, G. hirsutum, and P. strobus. In rice, AtCIPK19 enhanced cold stress tolerance by expression of OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH, OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4. To our best knowledge, this is the first report that describes a detailed interaction between AtCIPK19 and genes associated with biosynthesis of Put, Spd, and Spm in improving cold stress tolerance in different plant species.
Materials and Methods
Plasmid constructs
The cDNA sequence of AtCIPK19 was amplified from the Arabidopsis genome and cloned into expression vector pCAMBIA1301 as previously described [27]. DNA of pCAMBIA1301 and AtCIPK19 were digested by KpnI and BamHI (Promega, Madison, WI, USA) at 37oC. DNA was purified using QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA) and ligated to generate the expression vector pCAMBIA1301-AtCIPK19. Expression vectors were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The neomycin phosphotransferase II (NPT II) gene was used as a select marker in the transformation.
Transformation of O. sativa, G. hirsutum, and P. strobus cells
AtCIPK19 transgenic cell lines of O. sativa, G. hirsutum, and P. strobus were generated as described before [28], using Agrobacterium tumefaciens strain GV3101 carrying pCAMBIA1301-AtCIPK19 to transform cultured cells. After transformation, cell cultures of different cell lines were growing for 4-5 week, and then these cell cultures were used for further analysis including cold stress and gene expression.
Polymerase chain reaction analyses of transgenic cells
Polymerase chain reaction (PCR) analysis of transgenic cells of O. sativa, G. hirsutum, and P. strobus was conducted as previously described [28]. One gram of control cells and transgenic cells were used to isolate genomic DNA, using a Genomic DNA Isolation Kit (Sigma). The primers used to amplify the NPTII gene are forward primer (nfp) 5’-GTCGACATGGCGGAGGAATTTGGAAGCATAG-3’ and the reverse primer (nrp) 5’-CCATGGTAGACTCCTGCTTCGACATCATGG-3’. The primers used to amplify the AtCIPK19 gene are forward primer (ckr) 5’-CTTTCAATGG CGGATTTGTT AAGAAAAGTG -3’ and the reverse primer (ckf) 5’- CTTCATTTTT CATCTTCCTA ATCAGTATCA GAAAG- 3’. The PCR mixture, the PCR conditions, and gel electrophoresis were carried out as described previously [28].
Southern blot analysis of transgenic cells
Southern blotting analysis of transgenic cells of O. sativa, G. hirsutum, and P. strobus was conducted as previously described [28]. Five grams of control cells and transgenic cells were used to isolate genomic DNA, using a Genomic DNA Isolation Kit (Sigma). Twenty-five micrograms of DNA were digested 16 hours with the enzymes Xba I (Boehringer Mannheim) at 37oC. The molecular probes (1452 pb fragment of AtCIPK19) were labeled by Digoxigenin (DIG) (Roche Diagnostics, Indianapolis, IN, USA).
RNA isolation and Northern blot analysis
Five grams of fresh cultures of transgenic and control cells of O. sativa, G. hirsutum, and P. strobus were used to isolate total RNA, using a RNeasy Mini Plant Kit (Germantown, MD, USA) by following the manual. Six ?g of total RNA was separated by agarose-gel electrophoresis. Electrophoresis of RNAs and northern blotting were performed as described before [28]. The hybridization probe is the Digoxigenin (DIG)-labelling AtCIPk19 DNA (1452 pb) (Roche Diagnostics). The rRNA was used as the loading control of RNA samples.
Cold treatment and determination of cell viability and cell growth rate
Cold treatment was performed by incubation of AtCIPK19 transgenic cells (at the age of 4-5 weeks) and control cells of O. sativa, G. hirsutum, and P. strobus at -10, 4, 12, and 24oC in the dark for 24 hours in growth chambers (Beijing ZNYT, China). Control seedlings were grown under the same conditions. After 24 h of chilling treatment, cells were moved to the normal growth environment. The influences of cold stress on cell growth and cell viability of O. sativa, G. hirsutum, and P. strobus were determined, as previously described [29]. The average growth was expressed as mg/g FW/day. The rate of cell growth and the cell viability were measured 7 days after treatment. Samples from both chilled and control samples were collected with 3 biological replicates.
Measurement of thiobarbituric acid reactive substances (TBARS)
Lipid peroxidation was determined as the amount of thiobarbituric acid reactive substances (TBARS) measured by the thiobarbituric acid (TBA) reaction as described previously (Tang and Page 2013). Transgenic and control cells (1 g) were homogenized in 3 ml of 20 % (w/v) trichloroacetic acid (TCA), then centrifuged at 5,000 rpm for 20 min, following by mixing with 20 % TCA containing 0.5 % (w/v) TBA and100 ?l 4 % BHT in ethanol at 1:1. The absorbance of the extracts of different cell lines was measured at 532 nm. The value of TBARS was calculated using the method described previously [28,29].
Determination of polyamines
Determination of polyamines putrescine (Put), spermidine (Spd), and spermine (Spm) from tissues of P. strobus was carried out as described previously [30]. Samples were examined using a HPLC and Spector Monitor 3200 Detector by following the manual of the facility. The measured polyamines are total PAs.
Expression of genes associated with biosynthesis of putrescine, spermidine, and spermine
Expression of genes associated with biosynthesis of putrescine (Put), spermidine (Spd), and spermine (Spm) in different cell lines of O. sativa, G. hirsutum, and P. strobus was examined using qRT–PCR by the method of [16,31,32]. Genes associated with biosynthesis of Put, Spd, and Spm include OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH, OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4 in AtCIPK19 transgenic cell lines have examined. The U6 gene was used as internal reference of qRT-PCR because U6 is one of most suitable reference gene under salinity and cold stresses [16,31-34]. The qRT-PCR data were normalized to the U6 internal control. The regular qRT-PCR was used. The delta-delta Ct method was used to obtain expression value.
RNA isolation and qRT-PCR
Total RNA was isolated using the regents of Sepasol RNA I Super (Nacalai Tesque, Kyoto, Japan). To synthesize the first-strand cDNA in a 50-mL reaction containing 2.5 mM oligo(dT) primers, 2.5 mM random heximer, and 2.5 mg of total RNA, the PrimeScript™ RT reagent kit (TaKaRa Co., Ltd, Ohtsu, Japan) was used by following the instruction. The qRT–PCR was performed using 1Å~ SYBR Premix Ex Taq II (TaKaRa). Five microliters of each reaction mixture was used as a template for PCR amplification in a 25-mL mixture containing 1.5 mM MgCl2, 200 mM. The primer sequences used for genes of OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH, OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4 are as described previously (Do, et al. 2013). The U6 gene was used as internal reference [16,31-34]. The data were normalized to the internal control. The delta-delta Ct method was used to obtain expression value.
Statistical analyses
Statistical analysis was performed using the General Linear Model procedure of SAS (Cary, NC, USA), employing ANOVA models. The significant differences between mean values of different cell lines derived from O. sativa, G. hirsutum, and P. strobus were made at 5 % level of probability, using the T-test. Each value of different cell lines derived from O. sativa, G. hirsutum, and P. strobus was presented as means standard errors of the mean.
Results
Generation of transgenic cell lines in O. sativa, G. hirsutum, and P. strobus
AtCIPK19 transgenic cell lines of O. sativa, G. hirsutum, and P. strobus were generated using Agrobacterium tumefaciens strain GV3101 carrying pCAMBIA1301-AtCIPK19 (Figure 1A) to transform cultured cells. After transformation, a total of 29 transgenic cell lines of O. sativa, a total of 32 transgenic cell lines of G. hirsutum, and a total of 36 transgenic cell lines of P. strobus were produced. After transgenic cell lines of O. sativa, G. hirsutum, and P. strobus were confirmed by PCR (Figure 1B,C), Southern blotting (Figure 1D), and northern blotting analyses (Figure 1E), nine cell lines (Os1, Os2, and Os3 from O. sativa, Gh1, Gh2, and Gh3 from G. hirsutum, and Ps1, Ps2, and Ps3 from P. strobus) from each of O. sativa, G. hirsutum, and P. strobus were selected and analyzed in this study.
Figure 1: Expression vector and generation of transgenic cells.
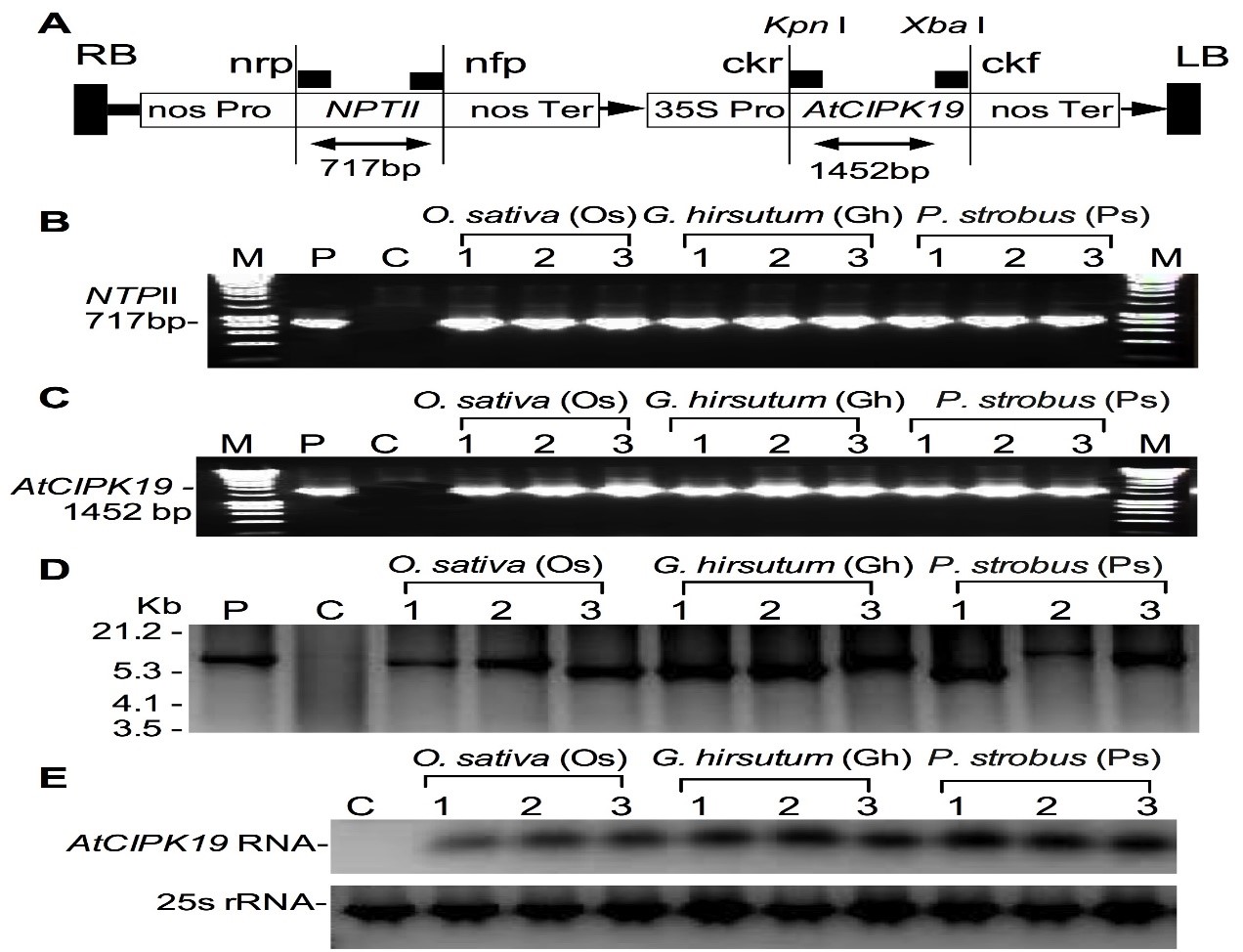
Overexpression of the AtCIPK19 gene increases the levels of putrescine (Put), spermidine (Spd), and spermine (Spm) in transgenic cells of O. sativa (A, B, and C), G. hirsutum (D, E, and F), and P. strobus (G, H, and I), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Overexpression of AtCIPK19 increases cell viability, cell growth rate, and decreases TBARS
To determine if overexpression of the AtCIPK19 gene increases cold stress tolerance, the cell growth rate, the cell viability, and the TBARS in transgenic cell lines of O. sativa, G. hirsutum, and P. strobus were examined. The cell growth rate and the cell viability were significantly increased in transgenic cell lines of O. sativa (Figure 2A,D), G. hirsutum (Figure 2B,E), and P. strobus (Figure 2C,F), respectively, under stress at -10oC and -4oC. The cell growth rate and the cell viability were not significantly increased in transgenic cell lines of O. sativa (Figure 2A,D), G. hirsutum (Figure 2B,E), and P. strobus (Figure 2C,F), respectively, under stress at 10oC and 24oC. The amount of TBARS was significantly increased in transgenic cell lines of O. sativa (Fig. 2G), G. hirsutum (Figure 2H), and P. strobus (Figure 2I), respectively, under stress at -10oC and -4oC. The amount of TBARS was not significantly increased in transgenic cell lines of O. sativa (Figure 2G), G. hirsutum (Fig. 2H), and P. strobus (Figure 2I), respectively, under stress at 10oC and 24oC.
Figure 2: Overexpression of AtCIPK19 increases cell viability, cell growth rate, and decreases TBARS.
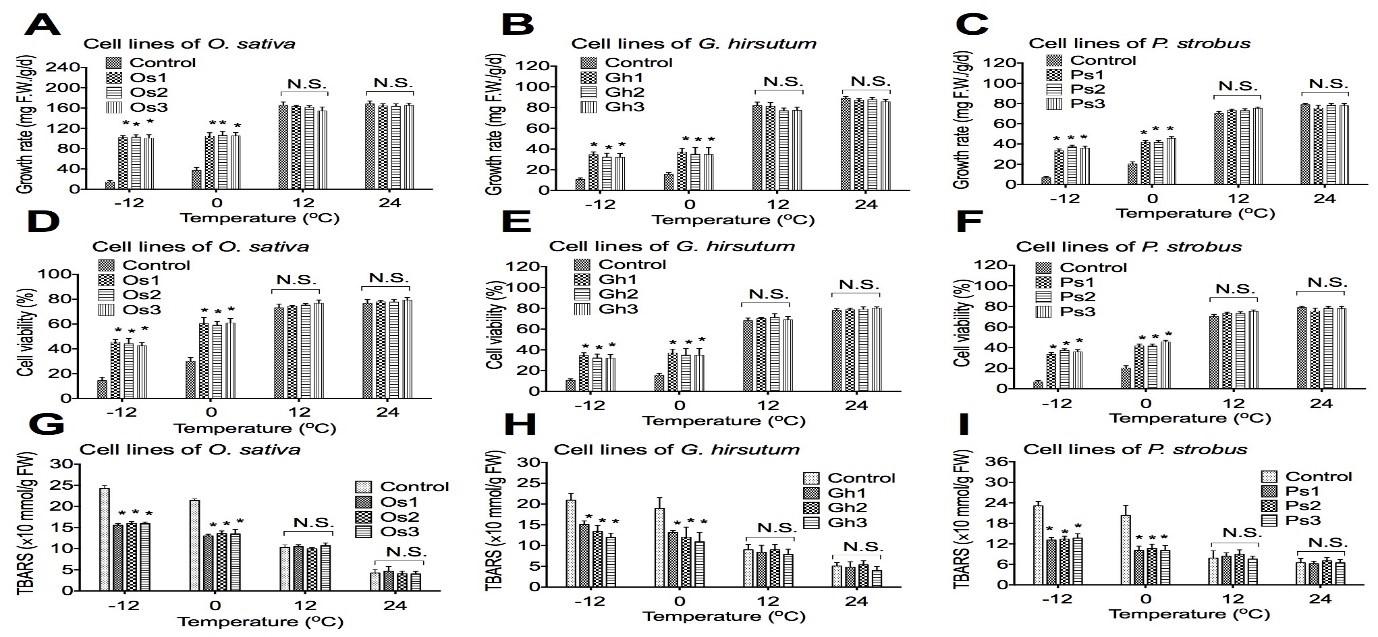
Overexpression of the AtCIPK19 gene increases the cell growth rate and the cell viability, but decreases the TBARS in transgenic cells of O. sativa (A, D, and G), G. hirsutum (B, E, and H), and P. strobus (C, F, and I), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard deviation. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Overexpression of AtCIPK19 increases contents of Put, Spd, and Spm
To examine if overexpression of the AtCIPK19 gene-enhanced cold stress tolerance is related to the biosynthesis of putrescine (Put), spermidine (Spd), and spermine (Spm), the contents of Put, Spd, and Spm were analyzed in transgenic cell lines of O. sativa, G. hirsutum, and P. strobus. Overexpression of the AtCIPK19 gene significantly increased the levels of putrescine (Put), spermidine (Spd), and spermine (Spm) in transgenic cells of O. sativa (Figure 3A,B and C), G. hirsutum (Figure 3D,E, and F), and P. strobus (Fig. 3G, H, and I), respectively, under stress at -10oC and -4oC. Overexpression of the AtCIPK19 gene did not significantly increase the levels of putrescine (Put), spermidine (Spd), and spermine (Spm) in transgenic cells of O. sativa (Fig. 3A, B, and C), G. hirsutum (Figure 3D,E, and F), and P. strobus (Figure 3G,H, and I), respectively, under stress at 10oC and 24oC.
Figure 3: Overexpression of AtCIPK19 increases contents of Put, Spd, and Spm.
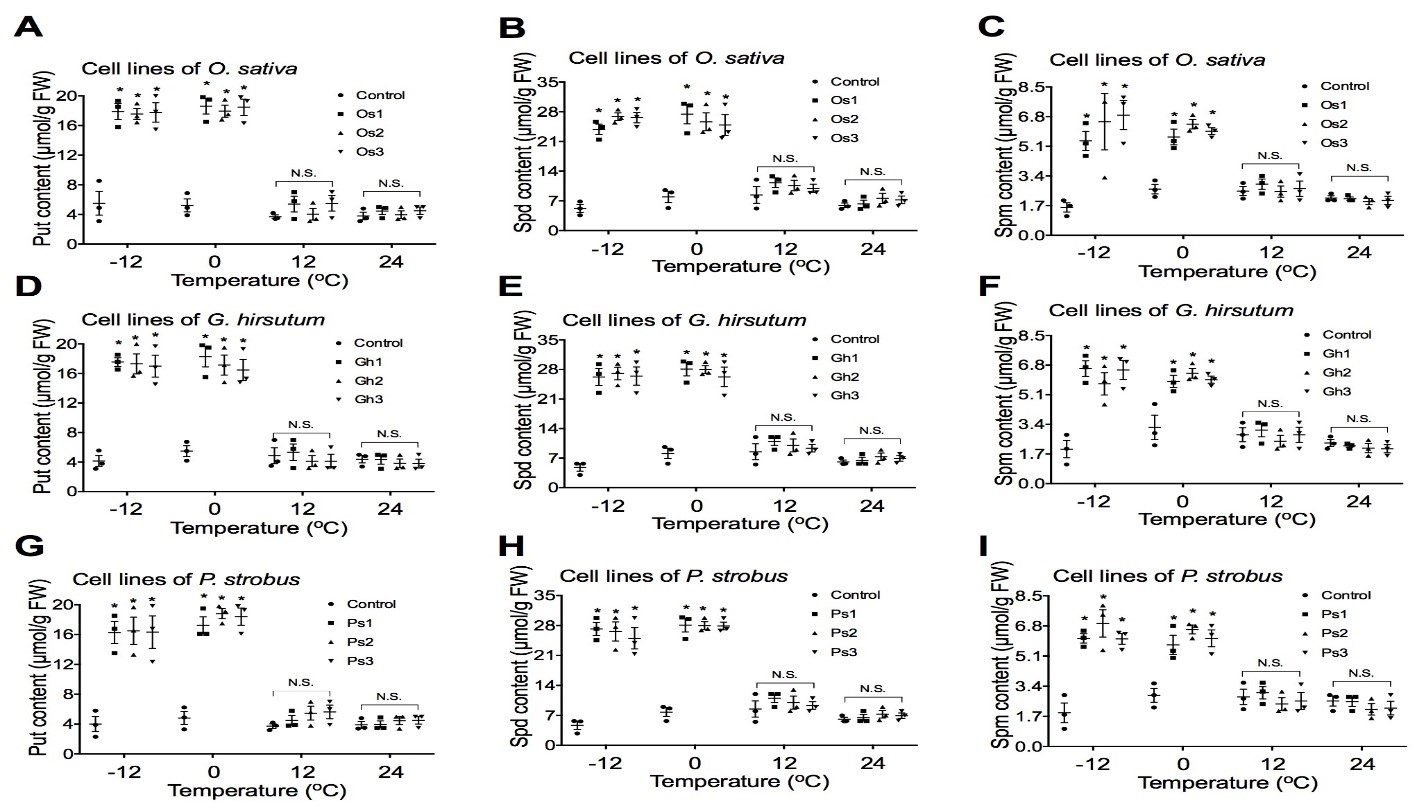
Overexpression of the AtCIPK19 gene increases the levels of putrescine (Put), spermidine (Spd), and spermine (Spm) in transgenic cells of O. sativa (A, B, and C), G. hirsutum (D, E, and F), and P. strobus (G, H, and I), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Expression of genes associated with putrescine biosynthesis in transgenic rice cells
To examine if overexpression of the AtCIPK19 gene-increased contents of Put, Spd, and Spm was related to the expression of genes associated with putrescine biosynthesis, expression of OsADC1 (Figure 4A), OsADC2 (Figure 4B), OsADC3 (Figure 4C), OsODC1 (Figure 4D), OsODC2 (Figure 4E), OsODC3 (Figure 4F), OsCPA1 (Figure 4G), OsCPA2 (Figure 4H), OsCPA3 (Figure 4I), and OsAIH (Figure 4J) were analyzed in transgenic cell lines (Os1, Os2, and Os3) of O. sativa. Overexpression of the AtCIPK19 gene significantly increases expression of putrescine biosynthesis enzymes genes (Figure 4) under stress at -10oC and -4oC. Overexpression of the AtCIPK19 gene does not significantly increases expression of putrescine biosynthesis enzymes genes (Figure 4) under stress at 10oC and 24oC.
Figure 4: Expression of genes associated with putrescine biosynthesis in transgenic rice cells.
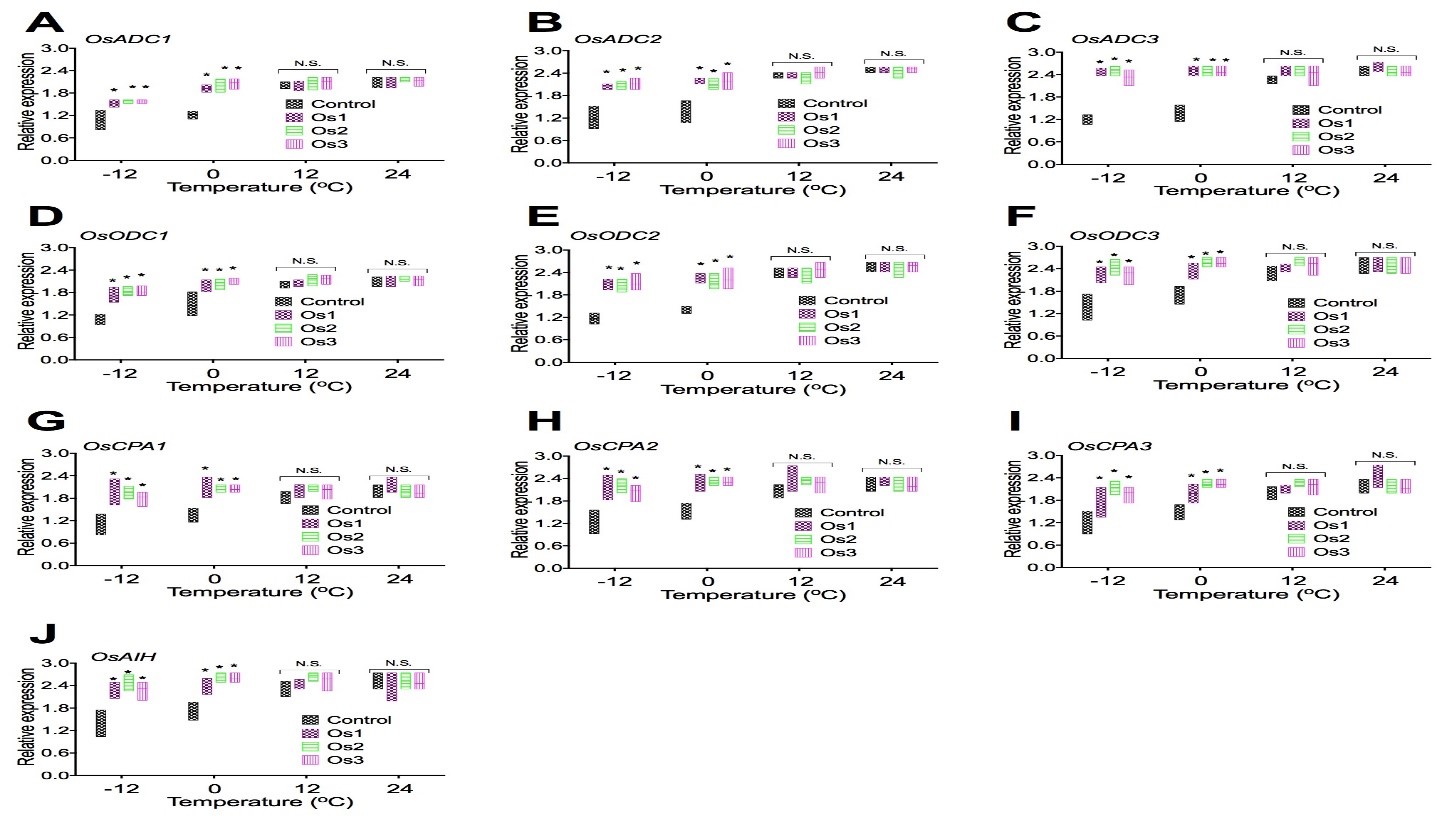
Overexpression of the AtCIPK19 gene increases expression of putrescine biosynthesis enzymes genes OsADC1 (A), OsADC2 (B), OsADC3 (C), OsODC1 (D), OsODC2 (E), OsODC3 (F), OsCPA1 (G), OsCPA2 (H), OsCPA3 (I), and OsAIH (J). The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Expression of genes associated with decarboxylated Sadenosylmethionine biosynthesis in transgenic rice cells
To examine if overexpression of the AtCIPK19 gene-increased contents of Put, Spd, and Spm was related to the expression of genes associated with decarboxylated Sadenosylmethionine biosynthesis, expression of OsSAMDC1 (Figure 5A), OsSAMDC2 (Figure 5B), OsSAMDC3 (Figure 5C), OsSAMDC4 (Figure 5D), OsSAMDC5 (Figure 5E), and OsSAMDC6 (Figure 5F) were analyzed in transgenic cell lines (Os1, Os2, and Os3) of O. sativa. Overexpression of the AtCIPK19 gene significantly increased expression of decarboxylated Sadenosylmethionine (dsSAM) biosynthesis enzymes genes OsSAMDC1 (Figure 5A), OsSAMDC2 (Figure 5B), OsSAMDC3 (Figure 5C), OsSAMDC4 (Figure 5D), OsSAMDC5 (Figure 5E), and OsSAMDC6 (Figure 5F) under stress at -10oC and -4oC. Overexpression of the AtCIPK19 gene did not significantly increase expression of decarboxylated Sadenosylmethionine (dsSAM) biosynthesis enzymes genes OsSAMDC1 (Figure 5A), OsSAMDC2 (Figure 5B), OsSAMDC3 (Figure 5C), OsSAMDC4 (Figure 5D), OsSAMDC5 (Figure 5E), and OsSAMDC6 (Figure 5F) under stress at 10oC and 24oC.
Figure 5: Expression of genes associated with decarboxylated Sadenosylmethionine biosynthesis in transgenic rice cells.
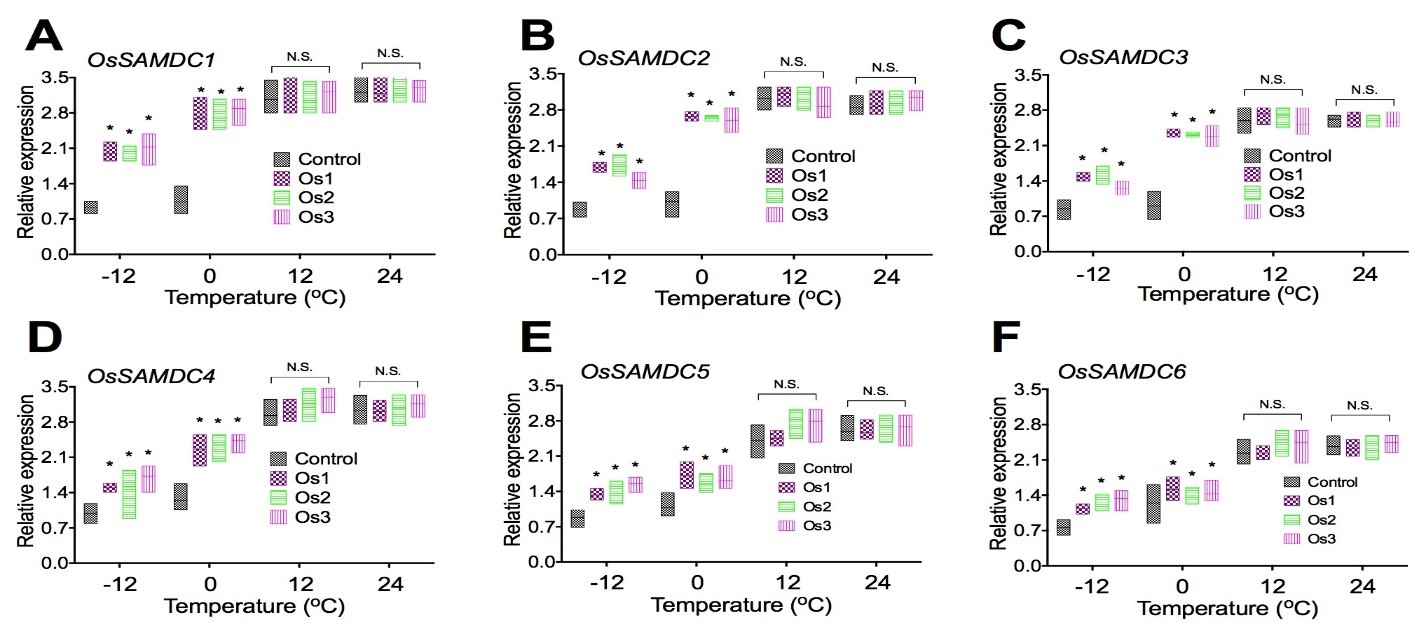
Overexpression of the AtCIPK19 gene increases expression of decarboxylated Sadenosylmethionine (dsSAM) biosynthesis enzymes genes OsSAMDC1 (A), OsSAMDC2 (B), OsSAMDC3 (C), OsSAMDC4 (D), OsSAMDC5 (E), and OsSAMDC6 (F). The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Expression of genes associated with spermidine and spermine biosynthesis in transgenic rice cells.
To examine if overexpression of the AtCIPK19 gene-increased contents of Put, Spd, and Spm was related to the expression of genes associated with spermidine and spermine biosynthesis, expression of OsSPD/SPM1 (Figure 6A), OsSPD/SPM2 (Figure 6B), OsSPD/SPM3 (Figure 6C), and OsSPD/SPM4 (Figure 6D) were analyzed in transgenic cell lines (Os1, Os2, and Os3) of O. sativa. Overexpression of the AtCIPK19 gene significantly increased expression of spermidine and spermine biosynthesis enzymes genes OsSPD/SPM1 (Figure 6A), OsSPD/SPM2 (Figure 6B), OsSPD/SPM3 (Figure 6C), and OsSPD/SPM4 (Figure 6D) under stress at -10oC and -4oC. Overexpression of the AtCIPK19 gene did not significantly increase expression of spermidine and spermine biosynthesis enzymes genes OsSPD/SPM1 (Figure 6A), OsSPD/SPM2 (Figure 6B), OsSPD/SPM3 (Figure 6C), and OsSPD/SPM4 (Figure 6D) under stress at 10oC and 24oC.
Figure 6: Expression of genes associated with spermidine and spermine biosynthesis in transgenic rice cells.
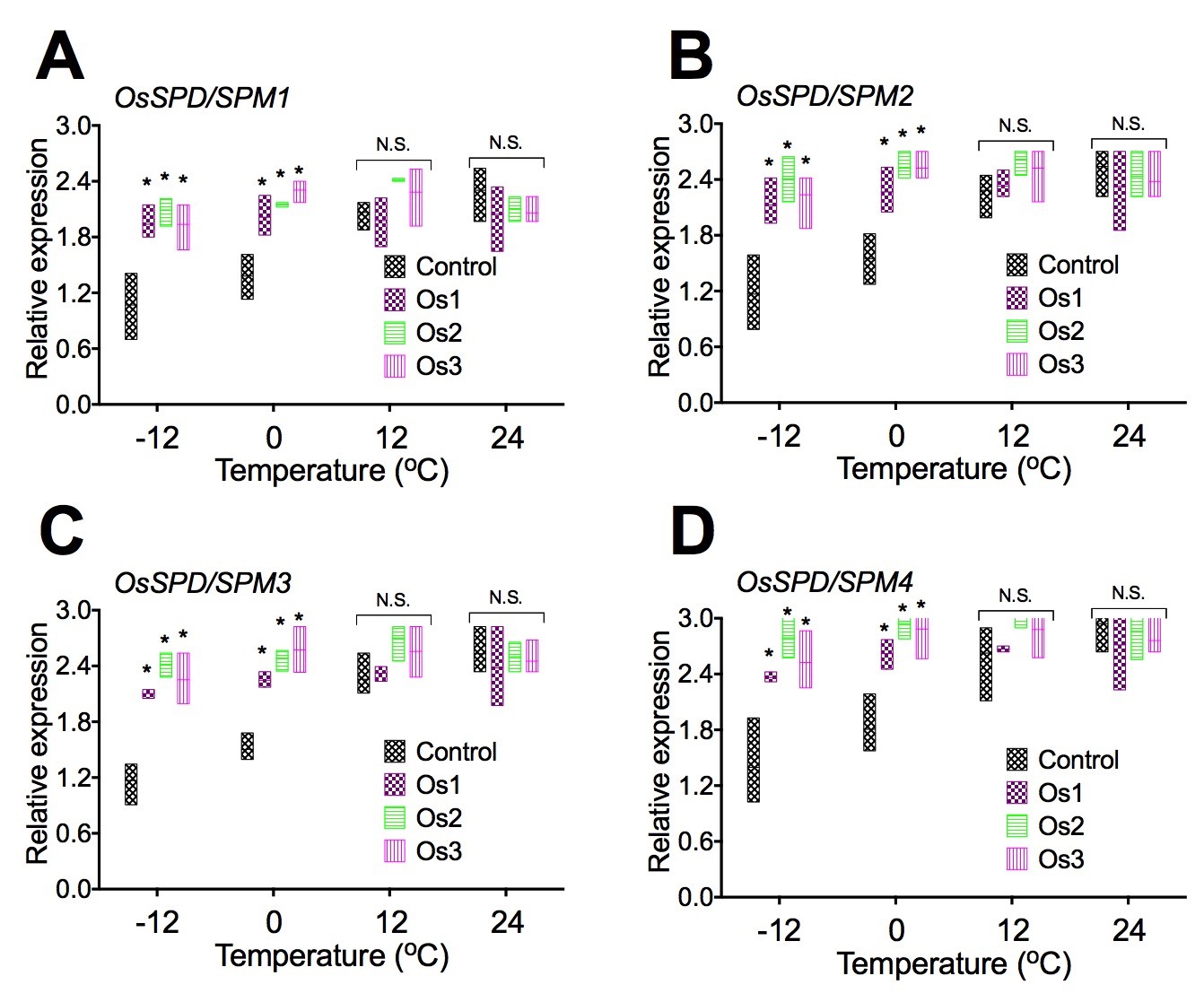
Overexpression of the AtCIPK19 gene increases expression of spermidine (Spd), and spermine (Spm) biosynthesis enzymes genes OsSPD/SPM1 (A), OsSPD/SPM2 (B), OsSPD/SPM3 (C), and OsSPD/SPM4 (D). The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, significant relative to control. N.S., no statistical significance.
Morphological changes of transgenic cell lines.
To examine if overexpression of the AtCIPK19 gene-increased contents of Put, Spd, and Spm caused morphological changes of transgenic cell lines, cell morphology was analyzed in transgenic cells in O. sativa (Figure 7A-D), G. hirsutum (Figure 6 7E-H), and P. strobus (Figure 7I-L), respectively, under treatment of -10oC -4oC, 10oC, and 24oC. Cold stress causes cell morphological change 3 days after treatment of different temperature at -10oC -4oC, 10oC, and 24oC.
Figure 7: Morphological changes of transgenic cell lines.
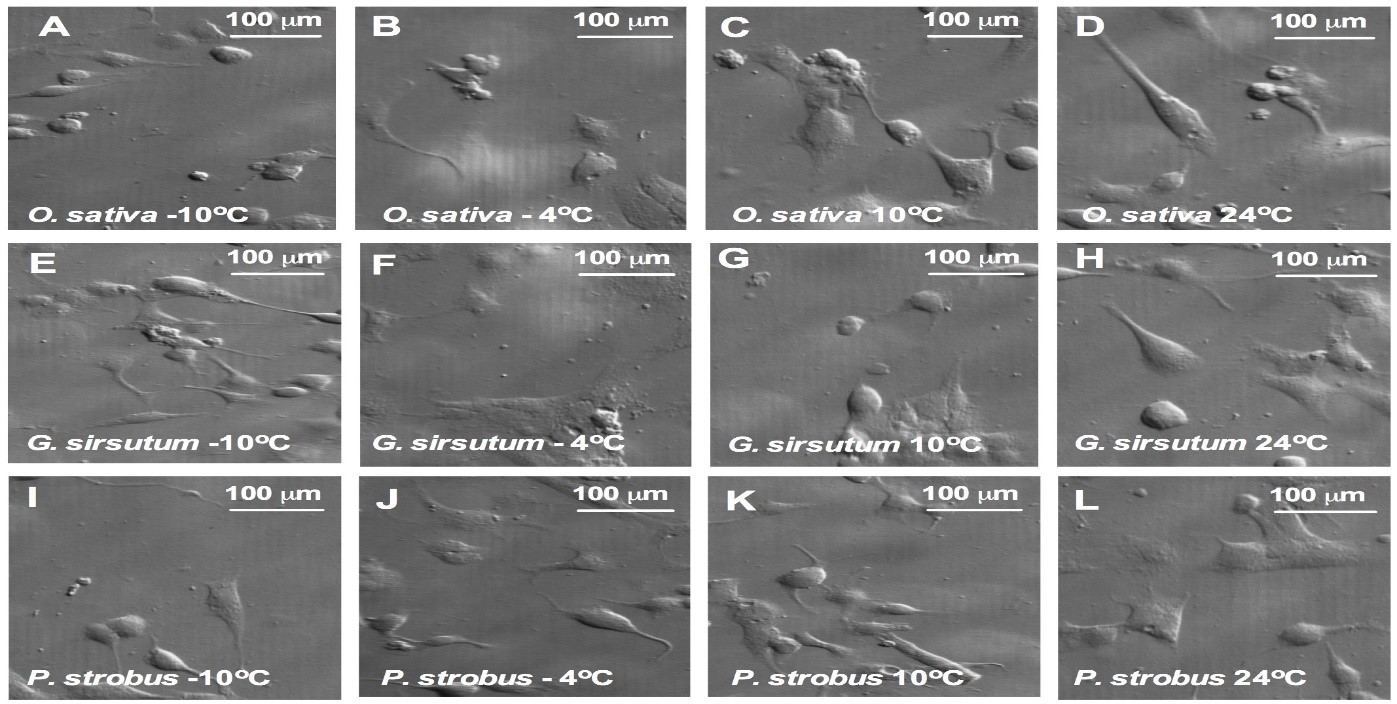
(a-d) Cell morphology of transgenic cells in O. sativa (A, B, C, and D), G. hirsutum (E, F, G, and H), and P. strobus (I, J, K, and L), respectively, under treatment of -10oC -4oC, 10oC, and 24oC. Cell images were taken 3 days after treatment of different temperature at -10oC -4oC, 10oC, and 24oC.
Discussion
Cold stress, which causes dehydration damage to the plant cell, is one of the most common abiotic stresses that adversely affect plant growth and crop productivity. Cold stress response is mediated by multiple signaling pathways and is regulated by many factors, among which CIPKs and polyamines may play a role. In Scots pine (Pinus sylvestris L.), temperature was a more effective signal than day-length for dehardening [36]. Phosphorylation-mediated signaling transduction plays a crucial role in the regulation of plant defense mechanisms against cold stress. In tomato, about 5500 phosphoproteins were identified to be involved in cold tolerant signaling [37]. In Carpobrotus edulis, cold stress reduced melatonin contents and increased salicylic acid contents [38]. GhHyPRP4 gene was isolated from cotton genome. Overexpression of GhHyPRP4 in yeast (Schizosaccharomyces pombe) significantly enhanced the cell survival rate upon treatment under -20 degrees C for 60 h, indicating that GhHyPRP4 may be involved in plant response to cold stress [39]. Expression of dehydrins is upregulated under cold stress, suggesting their involvement in membrane stabilization [40]. In Brassica napus, BnCOR25 was expressed at high levels in hypocotyls, cotyledons, stems, and flowers, and BnCOR25 transcripts were significantly induced by cold stress treatment [41].
The signal transduction of the plant hormone abscisic acid (ABA) has been studied extensively. ABA plays an important role in improving plant tolerance to cold via integrating sugars and reactive oxygen species signaling pathways [42-43]. Plants counteract cold stress through signal transduction and molecular genetic regulation. In our study, genetic and biochemical assays were performed to explore the effect of AtCIPK19 in response to cold stress in different plant species. Overexpression of AtCIPK19 increased the cell viability, the cell growth rate, and decreases the TBARS in cells of O. sativa, G. hirsutum, and P. strobus after treatment at -10, and 4oC, respectively. These results demonstrated that overexpression of AtCIPK19 enhanced cold stress tolerance could be achieved in different plant species [27,49,50].
Polyamines (PAs) are vital signals in modulating plant response to abiotic stress. Polyamines have been globally associated to plant responses to abiotic stress. Particularly, putrescine has been related to a better response to cold and dehydration stresses [51,52]. It is known that this polyamine is involved in cold tolerance. Cold temperature inhibits stomatal opening and causes stomatal closure. Cold-acclimated plants often exhibit marked changes in their lipid composition, particularly of the membranes. Cold stress often leads to the accumulation of ABA, glycine betaine, polyamines, and proline [51]. In Leymus chinensis, the S-adenosylmethionine decarboxylase gene, LcSAMDC1, was up-regulated by cold stress. Overexpression of LcSAMDC1 in transgenic Arabidopsis promoted increased tolerance to cold stress, indicating that LcSAMDC1 could be used to improve the abiotic resistance of crops [52]. Studies on the relationship between NO and PAs in response to cold stress in tomato showed that NO induced by Spd plays a crucial role in tomato's response to chilling stress [53]. In Arabidopsis, the level of putrescine increased substantially under cold stress [54]. In Arabidopsis thaliana, the increment in putrescine upon cold treatment correlated with the induction of known stress-responsive genes, and putrescine may be directly or indirectly involved in ABA metabolism and gene expression [55]. The levels of endogenous polyamines have been shown to increase in plant cells challenged with low temperature. The accumulation of putrescine under cold stress is essential for proper cold acclimation and survival at freezing temperatures [56-58]. Cold stress has been the subject of intense investigation to unravel the complex mechanisms responsible for cold tolerance. In this study, we foud that AtCIPK19 increases cold stress tolerance by regulating expression of OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH, OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4.
Calcineurin B-like proteins interacting protein kinases (CIPKs) play important roles in diverse plant stress responses. In Arabidopsis thaliana, CIPK21 ubiquitously expressed in tissues and up-regulated under multiple abiotic stress conditions. CIPK21 mediated responses to cold stress is associated with ion and water homeostasis [49]. In maize, ZmCIPK21 was primarily localized in the cytoplasm and the nucleus of cells and displayed enhanced expression under stress. Over-expression of ZmCIPK21 in wild-type Arabidopsis plants increased their tolerance to salt stress [59]. In Arabidopsis, overexpressing of CIPK9 resulted in a low-K(+)-sensitive phenotype [60]. In wheat, TaCIPK14 was upregulated under cold conditions. Transgenic tobaccos overexpressing TaCIPK14 exhibited higher contents of chlorophyll and sugar under cold stress [61]. In rice (Oryza sativa), OsCIPK14/15 play a crucial role in the microbe-associated molecular functions of stress tolerance [62]. Three CIPK genes (OsCIPK03, OsCIPK12, and OsCIPK15) were overexpressed in japonica rice and transgenic plants overexpressing the transgenes OsCIPK03, OsCIPK12, and OsCIPK15 showed significantly improved tolerance to cold stress [63]. In this investigation, our results demonstrated that AtCIPK19 increased cold stress tolerance by regulating expression of genes associated with putrescine biosynthesis including OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH.
Conclusion
In this investigation, we identified a mechanism of AtCIPK19 enhanced cold stress tolerance. We found that overexpression of AtCIPK19 increased cell viability and growth rate under cold stress. AtCIPK19 increased cold stress tolerance by regulating expression of genes associated with putrescine biosynthesis including OsADC1, OsADC2, OsADC3, OsODC1, OsODC2, OsODC3, OsCPA1, OsCPA2, OsCPA3, OsAIH. In AtCIPK19 transgenic rice cell lines, the transcript levels of OsSAMDC1, OsSAMDC2, OsSAMDC3, OsSAMDC4, OsSAMDC5, OsSAMDC6, OsSPD/SPM1, OsSPD/SPM2, OsSPD/SPM3, and OsSPD/SPM4 were increased, indicating that AtCIPK19 enhances cold stress tolerance by regulating expression of these genes in plant cells. These results will increase our understanding of AtCIPK19-related cold stress tolerance in different plant species including monocotyledonous, dicotyledonous, and gymnosperm plants.
Acknowledgement
We acknowledge the university central instrumentation facility for providing necessary experiment set up. We thank the University Council of Scientific Research for support. The authors are grateful to Dr. Bradshaw, Dr. Lischewski, and Dr. Page for their critical reading and suggestions during the preparation of this manuscript.
Author contributions
WT conceived and designed the experiments. WT wrote the paper. WT performed the experiment and analyzed the data. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Education Committee of Hubei Providence of China and by the National Natural Science Foundation of China (31270740).
References
- Nakaminami K, Seki M. 2018. RNA Regulation in Plant Cold Stress Response. Advances in experimental medicine and biology. 1081: 23-44.[Ref.]
- Zhuo X, Zheng T, Zhang Z, et al. 2018. Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes. 9.[Ref.]
- Zhou M, Tang W. 2018. MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Molecular genetics and genomics. MGG.[Ref.]
- Baumann K. 2017. Stress Responses: Membrane-to-nucleus signals modulate plant cold tolerance. Nature reviews Molecular cell biology. 18: 276-277.[Ref.]
- Zong JM, Li XW, Zhou YH, et al. 2016. The AaDREB1 Transcription Factor from the Cold-Tolerant Plant Adonis amurensis Enhances Abiotic Stress Tolerance in Transgenic Plant. International journal of molecular sciences. 17.[Ref.]
- Fei J, Wang YS, Zhou Q, et al. 2015. Cloning and expression analysis of HSP70 gene from mangrove plant Kandelia obovata under cold stress. Ecotoxicology. 24: 1677-1685.[Ref.]
- Kosova K, Vitamvas P, Prasil IT. 2014. Wheat and barley dehydrins under cold, drought, and salinity - what can LEA-II proteins tell us about plant stress response? Frontiers in plant science. 5: 343.[Ref.]
- Luo X, Bai X, Zhu D, et al. 2012. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta. 235: 1141-1155.[Ref.]
- Romero FM, Maiale SJ, Rossi FR, et al. 2018. Polyamine Metabolism Responses to Biotic and Abiotic Stress. Methods in molecular biology. 1694: 37-49.[Ref.]
- Gong X, Dou F, Cheng X, et al. 2018. Genome-wide identification of genes involved in polyamine biosynthesis and the role of exogenous polyamines in Malus hupehensis Rehd. under alkaline stress. Gene. 669: 52-62.[Ref.]
- Zhao M, Liu H, Deng Z, et al. 2017. Molecular cloning and characterization of S-adenosylmethionine decarboxylase gene in rubber tree (Hevea brasiliensis). Physiology and molecular biology of plants : an international journal of functional plant biology. 23: 281-290.[Ref.]
- Wang D, Li L, Xu Y, et al. 2017. Effect of Exogenous Nitro Oxide on Chilling Tolerance, Polyamine, Proline, and gamma-Aminobutyric Acid in Bamboo Shoots (Phyllostachys praecox f. prevernalis). Journal of agricultural and food chemistry. 65: 5607-5613.[Ref.]
- Zhang X, Wang W, Wang M, et al. 2016. The miR396b of Poncirus trifoliata Functions in Cold Tolerance by Regulating ACC Oxidase Gene Expression and Modulating Ethylene-Polyamine Homeostasis. Plant & cell physiology. 57: 1865-1878.[Ref.]
- Wang Y, Luo Z, Mao L, et al. 2016. Contribution of polyamines metabolism and GABA shunt to chilling tolerance induced by nitric oxide in cold-stored banana fruit. Food chemistry. 197: 333-339.[Ref.]
- Zhu X, Li Q, Hu J, et al. 2015. Molecular Cloning and Characterization of Spermine Synthesis Gene Associated with Cold Tolerance in Tea Plant (Camellia sinensis). Applied biochemistry and biotechnology. 177: 1055-1068.[Ref.]
- Basu S, Roychoudhury A, Sengupta DN. 2014. Identification of trans-acting factors regulating SamDC expression in Oryza sativa. Biochemical and biophysical research communications. 445: 398-403.[Ref.]
- Kovacs Z, Simon-Sarkadi L, Szucs A, et al. 2010. Differential effects of cold, osmotic stress and abscisic acid on polyamine accumulation in wheat. Amino acids. 38: 623-631.[Ref.]
- Alcazar R, Cuevas JC, Planas J, et al. 2011. Integration of polyamines in the cold acclimation response. Plant science : an international journal of experimental plant biology. 180: 31-38.[Ref.]
- Angelcheva L, Mishra Y, Antti H, et al. 2014. Metabolomic analysis of extreme freezing tolerance in Siberian spruce (Picea obovata). The New phytologist. 204: 545-555.[Ref.]
- Amarasinghe S, Watson-Haigh NS, Gilliham M, et al. 2016. The evolutionary origin of CIPK16: A gene involved in enhanced salt tolerance. Molecular phylogenetics and evolution. 100: 135-147.[Ref.]
- Beier MP, Obara M, Taniai A, et al. 2018. Lack of ACTPK1, an STY kinase, enhances ammonium uptake and use, and promotes growth of rice seedlings under sufficient external ammonium. The Plant journal : for cell and molecular biology. 93: 992-1006.[Ref.]
- Luo Q, Wei Q, Wang R, et al. 2017. BdCIPK31, a Calcineurin B-Like Protein-Interacting Protein Kinase, Regulates Plant Response to Drought and Salt Stress. Frontiers in plant science. 8: 1184.[Ref.]
- Rodriguez-Medina C, Boissinot S, Chapuis S, et al. 2015. A protein kinase binds the C-terminal domain of the readthrough protein of Turnip yellows virus and regulates virus accumulation. Virology. 486: 44-53.[Ref.]
- Deng X, Hu W, Wei S, et al. 2013. TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PloS one. 8: 69881.[Ref.]
- Kurusu T, Hamada J, Hamada H, et al. 2010. Roles of calcineurin B-like protein-interacting protein kinases in innate immunity in rice. Plant signaling & behavior. 5: 1045-1047.[Ref.]
- Pandey GK, Cheong YH, Kim BG, et al. 2007. CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell research. 17: 411-421.[Ref.]
- Zhou L, Lan W, Chen B, et al. 2015. A calcium sensor-regulated protein kinase, CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19, is required for pollen tube growth and polarity. Plant physiology. 167: 1351-1360.[Ref.]
- Tang W, Newton RJ, Li C, et al. 2007. Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant cell reports. 26: 115-124.[Ref.]
- Tang W, Page M. 2013. Transcription factor AtbZIP60 regulates expression of Ca2+ -dependent protein kinase genes in transgenic cells. Molecular biology reports. 40: 2723-2732.[Ref.]
- Tang W, Newton RJ. 2005. Polyamines promote root elongation and growth by increasing root cell division in regenerated Virginia pine (Pinus virginiana Mill.) plantlets. Plant cell reports. 24: 581-589.[Ref.]
- Do PT, Degenkolbe T, Erban A, et al. 2013. Dissecting rice polyamine metabolism under controlled long-term drought stress. PloS one. 8: 60325.[Ref.]
- Ding C, Chen T, Yang Y, et al. 2015. Molecular cloning and characterization of an S-adenosylmethionine synthetase gene from Chorispora bungeana. Gene. 572: 205-213.[Ref.]
- Wang M, Zhang X, Liu JH. 2015. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC genomics. 16: 555.[Ref.]
- Luo M, Gao Z, Li H, et al. 2018. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Scientific reports. 8: 4444.[Ref.]
- Yang Y, Zhang X, Chen Y, et al. 2016. Selection of Reference Genes for Normalization of MicroRNA Expression by RT-qPCR in Sugarcane Buds under Cold Stress. Frontiers in plant science. 7: 86.[Ref.]
- Beck EH, Heim R, Hansen J. 2004. Plant resistance to cold stress: mechanisms and environmental signals triggering frost hardening and dehardening. Journal of biosciences. 29: 449-459.[Ref.]
- Hsu CC, Zhu Y, Arrington JV, et al. 2018. Universal Plant Phosphoproteomics Workflow and Its Application to Tomato Signaling in Response to Cold Stress. Molecular & cellular proteomics : MCP. 17: 2068-2080.[Ref.]
- Fenollosa E, Gamez A, Munne-Bosch S. 2018. Plasticity in the hormonal response to cold stress in the invasive plant Carpobrotus edulis. Journal of plant physiology. 231: 202-209.[Ref.]
- Huang G, Gong S, Xu W, et al. 2011. GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta biochimica et biophysica Sinica. 43: 519-527.[Ref.]
- Eriksson SK, Kutzer M, Procek J, et al. 2011. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. The Plant cell. 23: 2391-2404.[Ref.]
- Chen L, Zhong H, Ren F, et al. 2011. A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant cell reports. 30: 463-471.[Ref.]
- Calegario FF, Cosso RG, Fagian MM, et al. 2003. Stimulation of potato tuber respiration by cold stress is associated with an increased capacity of both plant uncoupling mitochondrial protein (PUMP) and alternative oxidase. Journal of bioenergetics and biomembranes. 35: 211-220.[Ref.]
- Dong CH, Zolman BK, Bartel B, et al. 2009. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Molecular plant. 2: 59-72.[Ref.]
- Ostroumova EA, Ostroumov VA, Sumina ON, et al. 2001. The search for proteins with immunochemical affinity to plant stress proteins at cold-adapted endemic Baikal fishes. Journal of thermal biology. 26: 209-214.[Ref.]
- Sangwan V, Orvar BL, Beyerly J, et al. 2002. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. The Plant journal : for cell and molecular biology. 31: 629-638.[Ref.]
- Streb P, Aubert S, Gout E, et al. 2003. Reversibility of cold- and light-stress tolerance and accompanying changes of metabolite and antioxidant levels in the two high mountain plant species Soldanella alpina and Ranunculus glacialis. Journal of experimental botany. 54: 405-418.[Ref.]
- Xue-Xuan X, Hong-Bo S, Yuan-Yuan M, et al. 2010. Biotechnological implications from abscisic acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Critical reviews in biotechnology. 30: 222-230.[Ref.]
- Yamazaki T, Kawamura Y, Uemura M. 2009. Extracellular freezing-induced mechanical stress and surface area regulation on the plasma membrane in cold-acclimated plant cells. Plant signaling & behavior. 4: 231-233.[Ref.]
- Pandey GK, Cheong YH, Kim BG, et al. 2007. CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell research. 17: 411-421.[Ref.]
- Pandey GK, Kanwar P, Singh A, et al. 2015. Calcineurin B-Like Protein-Interacting Protein Kinase CIPK21 Regulates Osmotic and Salt Stress Responses in Arabidopsis. Plant physiology. 169: 780-792.[Ref.]
- Agurla S, Gahir S, Munemasa S, et al. 2018. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. Advances in experimental medicine and biology. 1081: 215-232.[Ref.]
- Liu Z, Liu P, Qi D, et al. 2017. Enhancement of cold and salt tolerance of Arabidopsis by transgenic expression of the S-adenosylmethionine decarboxylase gene from Leymus chinensis. Journal of plant physiology. 211: 90-99.[Ref.]
- Diao QN, Song YJ, Shi DM, et al. 2016. Nitric oxide induced by polyamines involves antioxidant systems against chilling stress in tomato (Lycopersicon esculentum Mill.) seedling. Journal of Zhejiang University Science B. 17: 916-930.[Ref.]
- Majlath I, Szalai G, Papp I, et al. 2011. Atnoa1 mutant Arabidopsis plants induce compensation mechanisms to reduce the negative effects of the mutation. Journal of plant physiology. 168: 1184-1190.[Ref.]
- Alet AI, Sanchez DH, Cuevas JC, et al. 2011. Putrescine accumulation in Arabidopsis thaliana transgenic lines enhances tolerance to dehydration and freezing stress. Plant signaling & behavior. 6: 278-286.[Ref.]
- Cuevas JC, Lopez-Cobollo R, Alcazar R, et al. 2008. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant physiology. 148: 1094-1105.[Ref.]
- Pillai MA, Akiyama T. 2004. Differential expression of an S-adenosyl-L-methionine decarboxylase gene involved in polyamine biosynthesis under low temperature stress in japonica and indica rice genotypes. Molecular genetics and genomics : MGG. 271: 141-149.[Ref.]
- Sfakianaki M, Sfichi L, Kotzabasis K. 2006. The involvement of LHCII-associated polyamines in the response of the photosynthetic apparatus to low temperature. Journal of photochemistry and photobiology B, Biology. 84: 181-188.[Ref.]
- Chen X, Huang Q, Zhang F, et al. 2014. ZmCIPK21, a maize CBL-interacting kinase, enhances salt stress tolerance in Arabidopsis thaliana. International journal of molecular sciences. 15: 14819-14834.[Ref.]
- Liu LL, Ren HM, Chen LQ, et al. 2013. A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant physiology. 161: 266-277.[Ref.]
- Deng X, Zhou S, Hu W, et al. 2013. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiologia plantarum. 149: 367-377.[Ref.]
- Kurusu T, Hamada J, Nokajima H, et al. 2010. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant physiology. 153: 678-692.[Ref.]
- Xiang Y, Huang Y, Xiong L. 2007. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant physiology. 144: 1416-1428.[Ref.]




















